From Your Question
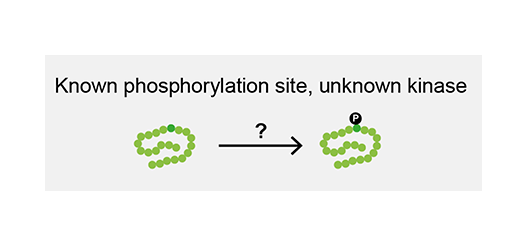
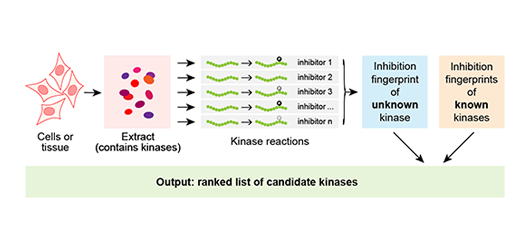
A phosphorylation site regulates the activity of your protein of interest. However, the kinase responsible is unknown. In silico methods might provide clues, but they do not account for cellular context and are often unreliable. Genetic screens can be unwieldy and often identify kinases that are only indirectly involved in phosphorylation. You need a direct experimental approach to address this question.